Session 4: loglinear regression part 1
Levi Waldron
session_lecture.RmdLearning objectives and outline
Learning objectives
- Define log-linear models in GLM framework
- Identify situations that motivate use of log-linear models
- Define the Poisson distribution and the log-linear Poisson GLM
- Identify applications and properties of the Poisson distribution
- Define multicollinearity and identify resulting issues
Brief review of GLMs
Components of GLM
- Random component specifies the conditional distribution for the response variable - it doesn’t have to be normal but can be any distribution that belongs to the “exponential” family of distributions
- Systematic component specifies linear function of predictors (linear predictor)
- Link [denoted by g(.)] specifies the relationship between the expected value of the random component and the systematic component, can be linear or nonlinear
Linear Regression as GLM
The model:
Random component of is normally distributed:
Systematic component (linear predictor):
Link function here is the identity link: . We are modeling the mean directly, no transformation.
Motivating example for log-linear models
Effectiveness of a depression case-management program
- Research question: can a new treatment reduce the number of needed visits to the emergency room, compared to standard care?
- outcome: # of emergency room visits for each patient in the year following initial treatment
-
predictors:
- race (white or nonwhite)
- treatment (treated or control)
- amount of alcohol consumption (numerical measure)
- drug use (numerical measure)
Statistical issues
- about 1/3 of observations are exactly 0 (did not return to the emergency room within the year)
- highly nonnormal and cannot be transformed to be approximately normal
- even transformation will have a “lump” at zero + over 1/2 the transformed data would have values of 0 or
- a linear regression model would give negative predictions for some covariate combinations
- some subjects die or cannot be followed up on for a whole year
Poisson log-linear GLM
Towards a reasonable model
- A multiplicative model will allow us to make inference on ratios of mean emergency room usage
- Modeling of the mean emergency usage ensures positive means, and does not suffer from problem
- Random component of GLM, or residuals (was for linear regression) may still not be normal, but we can choose from other distributions
Proposed model without time
Or equivalently: where is the expected number of emergency room visits for patient i.
- Important note: Modeling is not equivalent to modeling
Accounting for follow-up time
Instead, model mean count per unit time:
Or equivalently:
- is not a covariate, it is called an offset
The Poisson distribution
- Count data are often modeled as Poisson distributed:
- mean is greater than 0
- variance is also
- Probability density
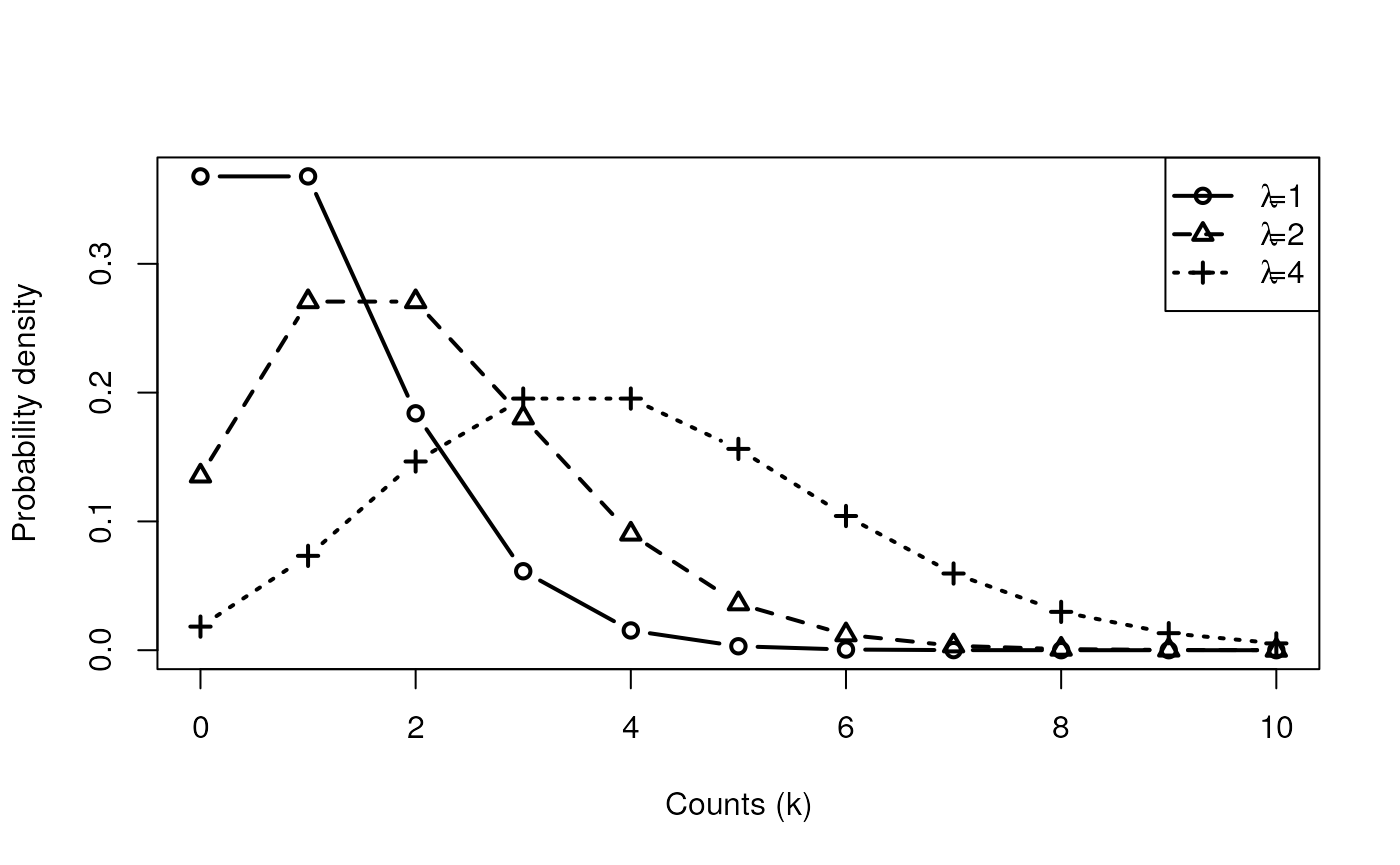
When the Poisson distribution works
- Individual events are low-probability (small p), but many
opportunities (large n)
- e.g. # 911 calls per day
- e.g. # emergency room visits
- Approximates the binomial distribution when n is large and p is
small
- e.g. , or
- When mean of residuals is approx. equal to variance
GLM with log-linear link and Poisson error model
- Model the number of counts per unit time as Poisson-distributed + so the expected number of counts per time is
Recalling the log-linear model systematic component:
GLM with log-linear link and Poisson error model (cont’d)
Then the systematic part of the GLM is: Or alternatively:
Interpretation of coefficients
- Suppose that in the fitted model, where for white and for non-white.
- The mean rate of emergency room visits per unit time for white relative to non-white, all else held equal, is estimated to be:
Interpretation of coefficients (cont’d)
- If
with whites as the reference group:
- after adjustment for treatment group, alcohol and drug usage, whites tend to use the emergency room at a rate 1.65 times higher than non-whites.
- equivalently, the average rate of usage for whites is 65% higher than that for non-whites
- Multiplicative rules apply for other coefficients as well, because they are exponentiated to estimate the mean rate.
Multi-collinearity
What is Multicollinearity?
- Multicollinearity exists when two or more of the independent variables in regression are moderately or highly correlated.
- High correlation among continuous predictors or high concordance among categorical predictors
- Impacts the ability to estimate regression coefficients
- larger standard errors for regression coefficients
- ie, coefficients are unstable over repeated sampling
- exact collinearity produces infinite standard errors on coefficients
- Can also result in unstable (high variance) prediction models
Identifying multicollinearity
- Pairwise correlations of data or of model matrix (latter works with categorical variables)
- Heat maps
- Variance Inflation Factor (VIF) of regression coefficients
Example: US Judge Ratings dataset
See ?USJudgeRatings for dataset, ?pairs for
plot code:
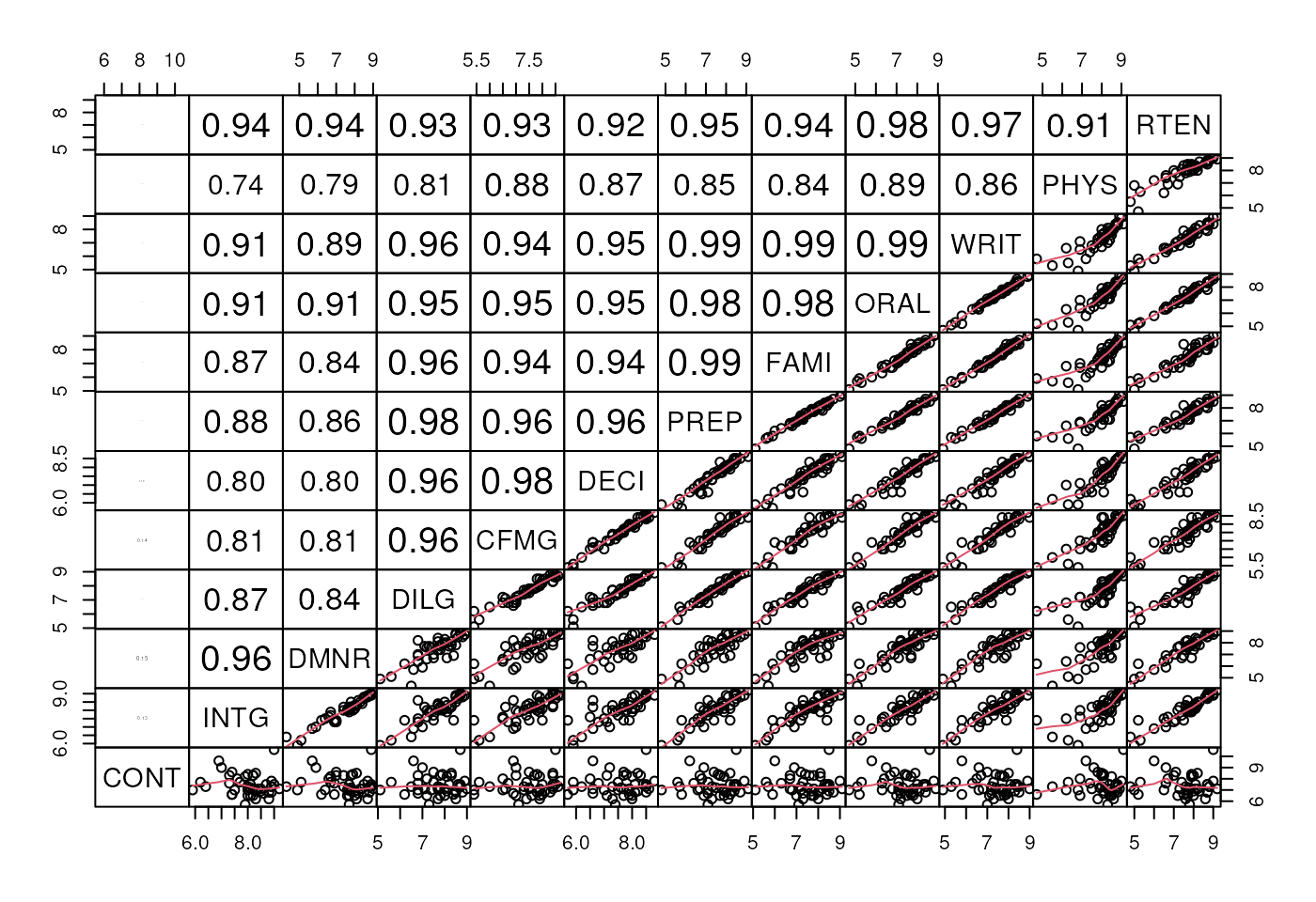 **Pairwise scatterplot of continuous variables in US Judge Ratings
dataset
**Pairwise scatterplot of continuous variables in US Judge Ratings
dataset
Example: iris dataset
One categorical variable, so use model matrix. Make a simple heatmap.
mm <- model.matrix( ~ ., data = iris)
pheatmap::pheatmap(cor(mm[, -1]), #-1 gets rid of intercept column
color = colorRampPalette(c("#f0f0f0", "#bdbdbd", "#636363"))(100))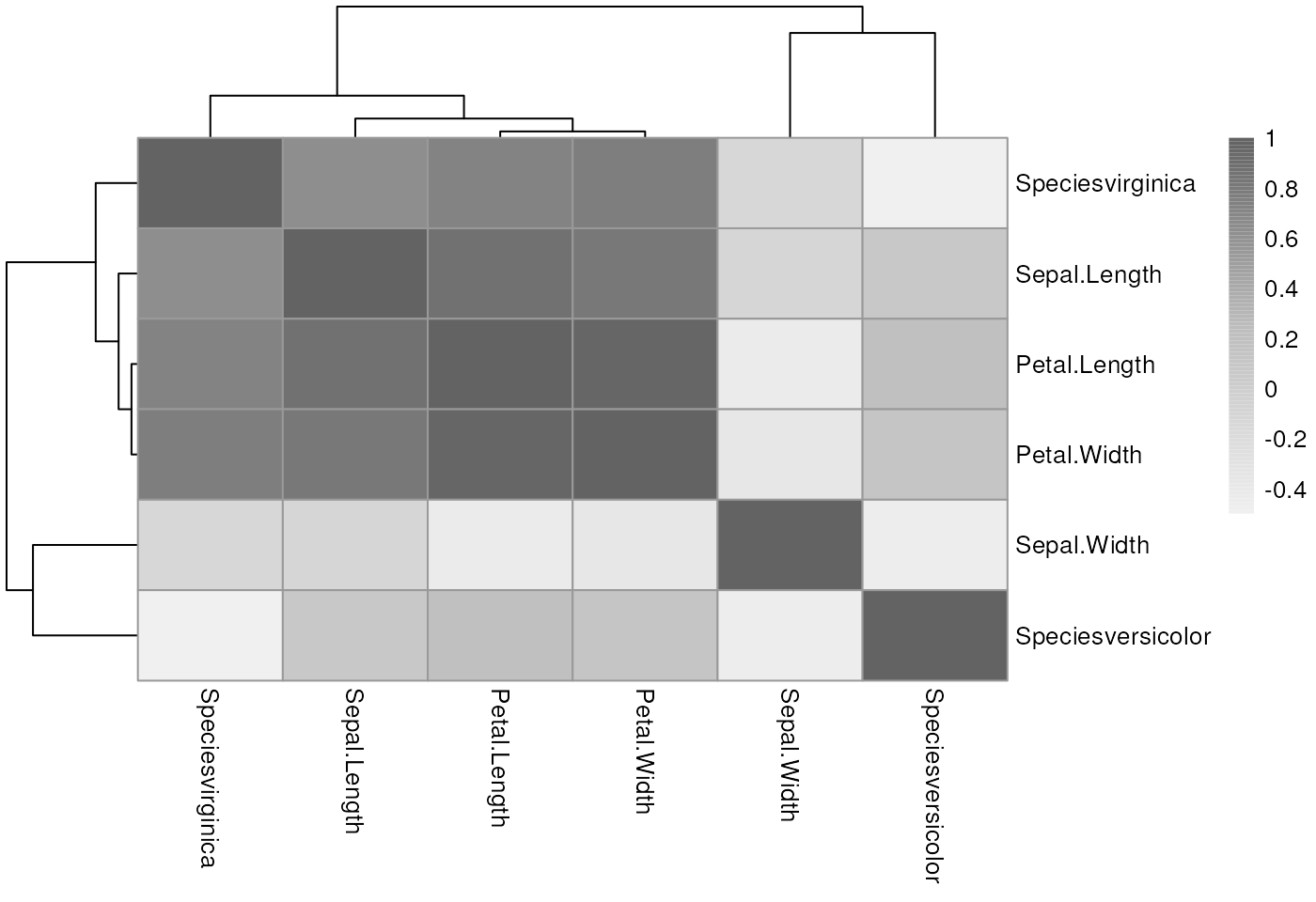 Note: multicollinearity exists between multiple predictors, not
between predictor and outcome
Note: multicollinearity exists between multiple predictors, not
between predictor and outcome
Conclusions
Conclusions
- Log-linear models are appropriate for non-negative, skewed count
data
- probability of each event is low
- The coefficients of log-linear models are multiplicative
- An offset term can account for varying follow-up time or otherwise varying opportunity to be counted
- Poisson distribution is limit of binomial distribution with high number of trials, low probability
- Inference from log-linear models is sensitive to the choice of error model (assumption on the distribution of residuals)
- We will cover other options next week for when the Poisson error
model doesn’t fit:
- Variance proportional to mean, instead of equal
- Negative Binomial
- Zero Inflation