Session 8: Survival analysis part 3
Levi Waldron
session_lecture.RmdLearning objectives and outline
Learning objectives
- Check model assumptions and fit of the Cox model
- residuals analysis
- log-minus-log plot
- Fit and interpret multivariate Cox models
- perform tests for trend
- predict survival for specific covariate patterns
- predict survival for adjusted coefficients
- Explain stratified analysis
- Identify situations of competing risks
- Describe the application of Propensity Score analysis
- Vittinghoff sections 6.2-6.4
Review
Cox proportional hazards model
- Cox proportional hazard regression assesses the relationship between
a right-censored, time-to-event outcome and multiple predictors:
- categorical variables (e.g., treatment groups)
- continuous variables
is the hazard of patient relative to baseline
is the time-dependent hazard function for patient
is the baseline hazard function, and is the negative of the slope of the , the baseline survival function.
Multiplicative model
Caveats and Assumptions
- Categories with no events
- can occur when the group is small or its risk is low
- HRs with respect to such a reference group are infinite
- hypothesis tests and CIs are difficult / impossible to interpret
- Assumptions of Cox PH model
- Constant hazard ratio over time (proportional hazards)
- Linear association between log(HR) and predictors (log-linearity) / multiplicative relationship between hazard and predictors
- Independence of survival times between individuals in the sample
- Uninformative censoring: a censored participant is the same as an uncensored participant with the same covariates at still in the risk set after that time
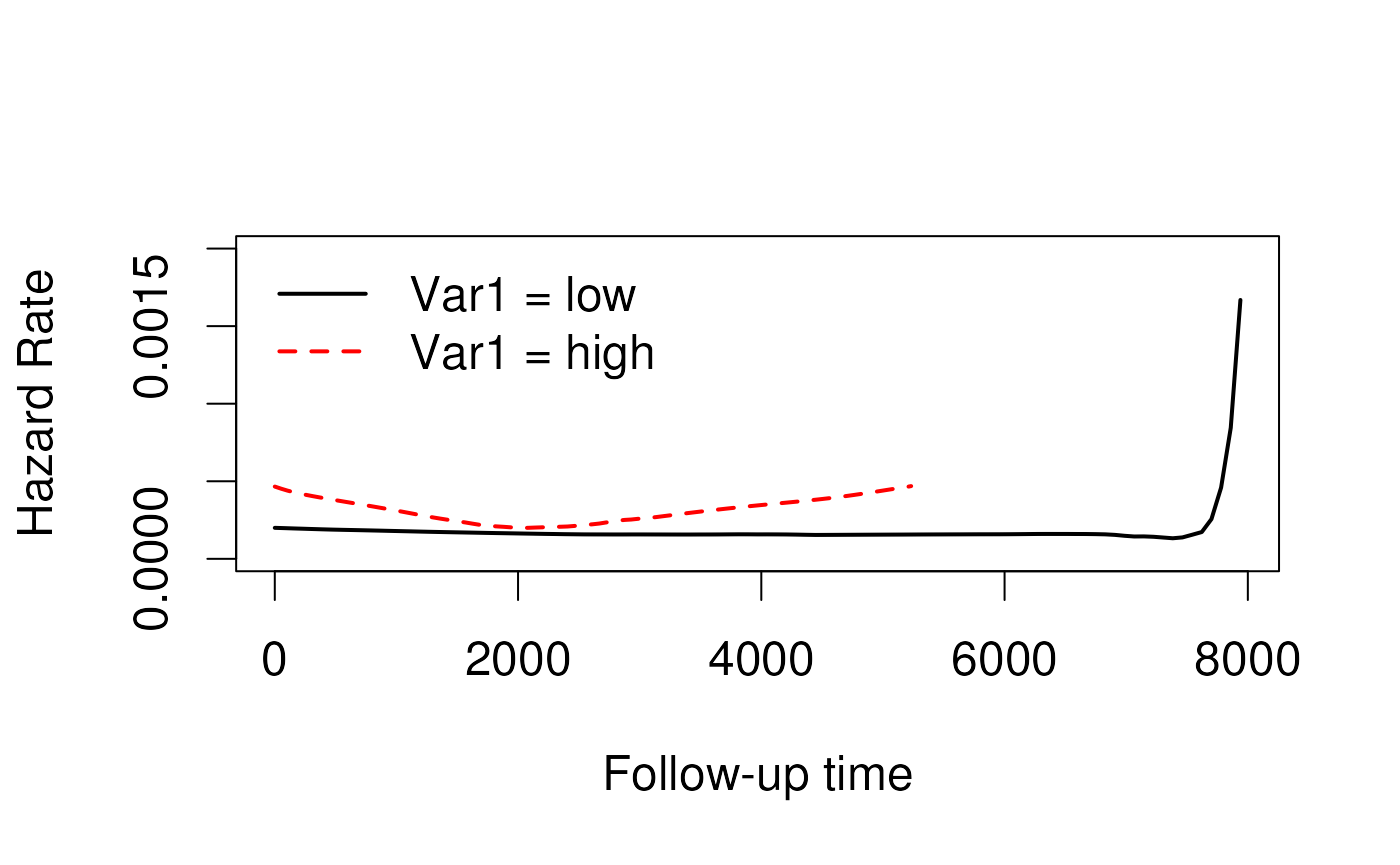
Checking assumptions of Cox model
Residuals analysis
- Residuals are used to investigate the lack of fit of a model to a given subject.
- For Cox regression, there’s no easy analog to the usual “observed minus predicted” residual
suppressPackageStartupMessages(library(pensim))
set.seed(1)
mydat <- create.data(
nvars = c(1, 1),
nsamples = 500,
cors = c(0, 0),
associations = c(0.5, 0.5),
firstonly = c(TRUE, TRUE),
censoring = c(0, 8.5)
)$dataRename variables of simulated data, and make one variable categorical:
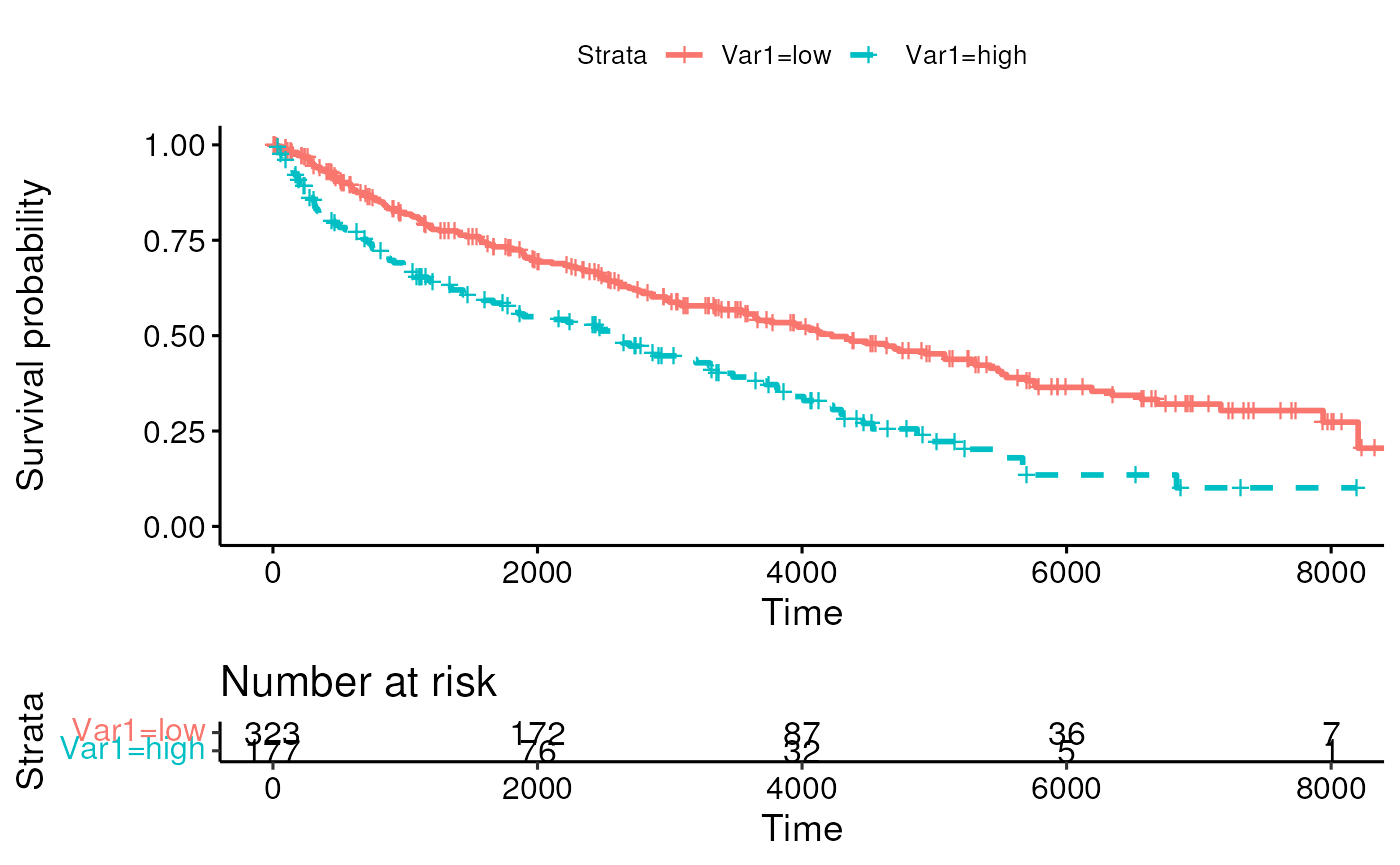
Simulated data to test residuals methods
summary(mydat)## Var1 Var2 time cens
## low :323 Min. :-2.99695 Min. : 5 Min. :0.000
## high:177 1st Qu.:-0.79008 1st Qu.: 691 1st Qu.:0.000
## Median :-0.02126 Median :1970 Median :1.000
## Mean :-0.04594 Mean :2529 Mean :0.526
## 3rd Qu.: 0.68933 3rd Qu.:3874 3rd Qu.:1.000
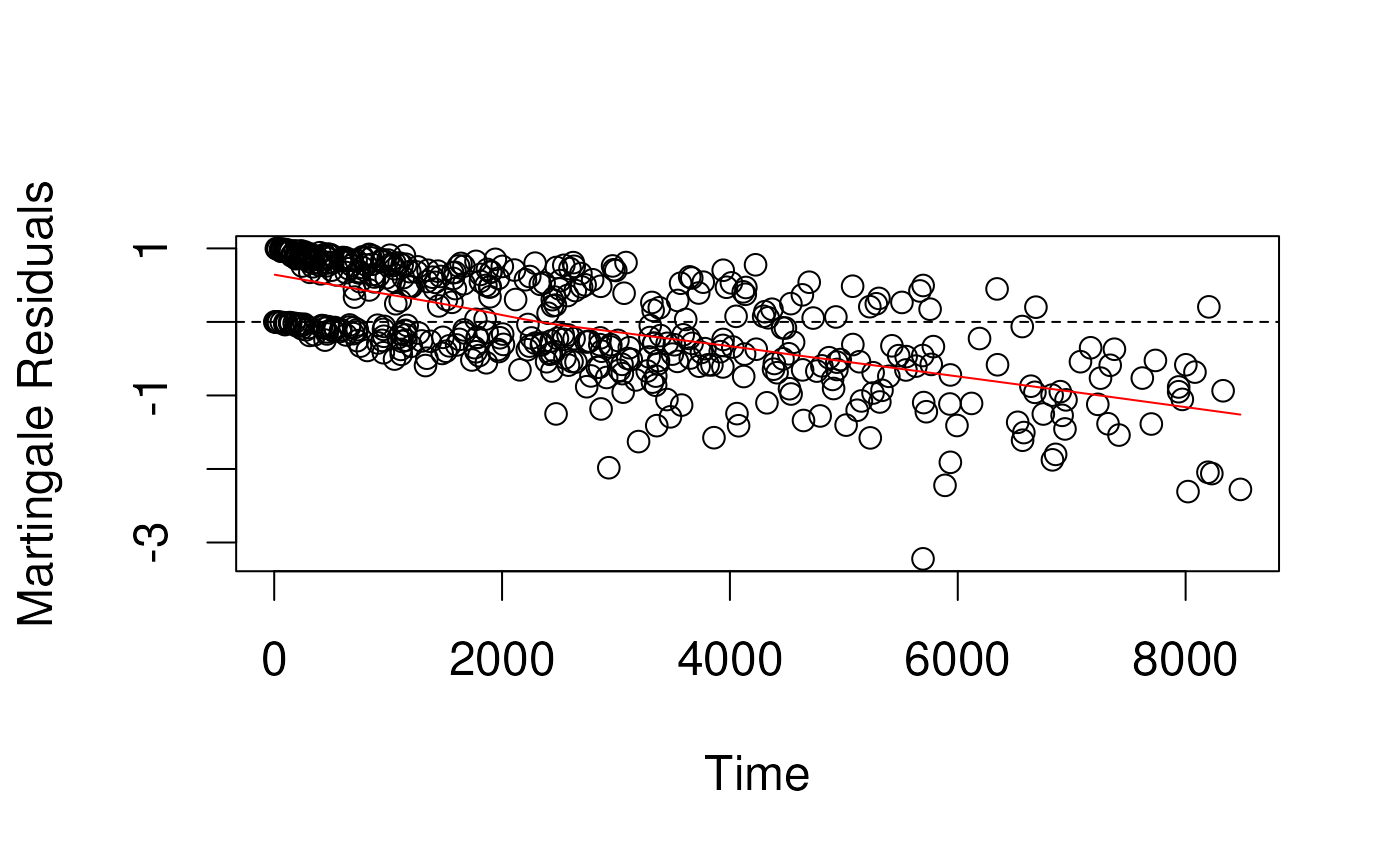
## Max. : 3.05574 Max. :8481 Max. :1.000Martingale residuals
- censoring variable
(1 if event, 0 if censored) minus the estimated cumulative hazard
function
(1 - survival function)
- E.g., for a subject censored at 1 year (), whose predicted cumulative hazard at 1 year was 30%, Martingale = .
- E.g. for a subject who had an event at 6 months, and whose predicted cumulative hazard at 6 months was 80%, Margingale = .
- Problem: not symmetrically distributed, even when model fits the data well
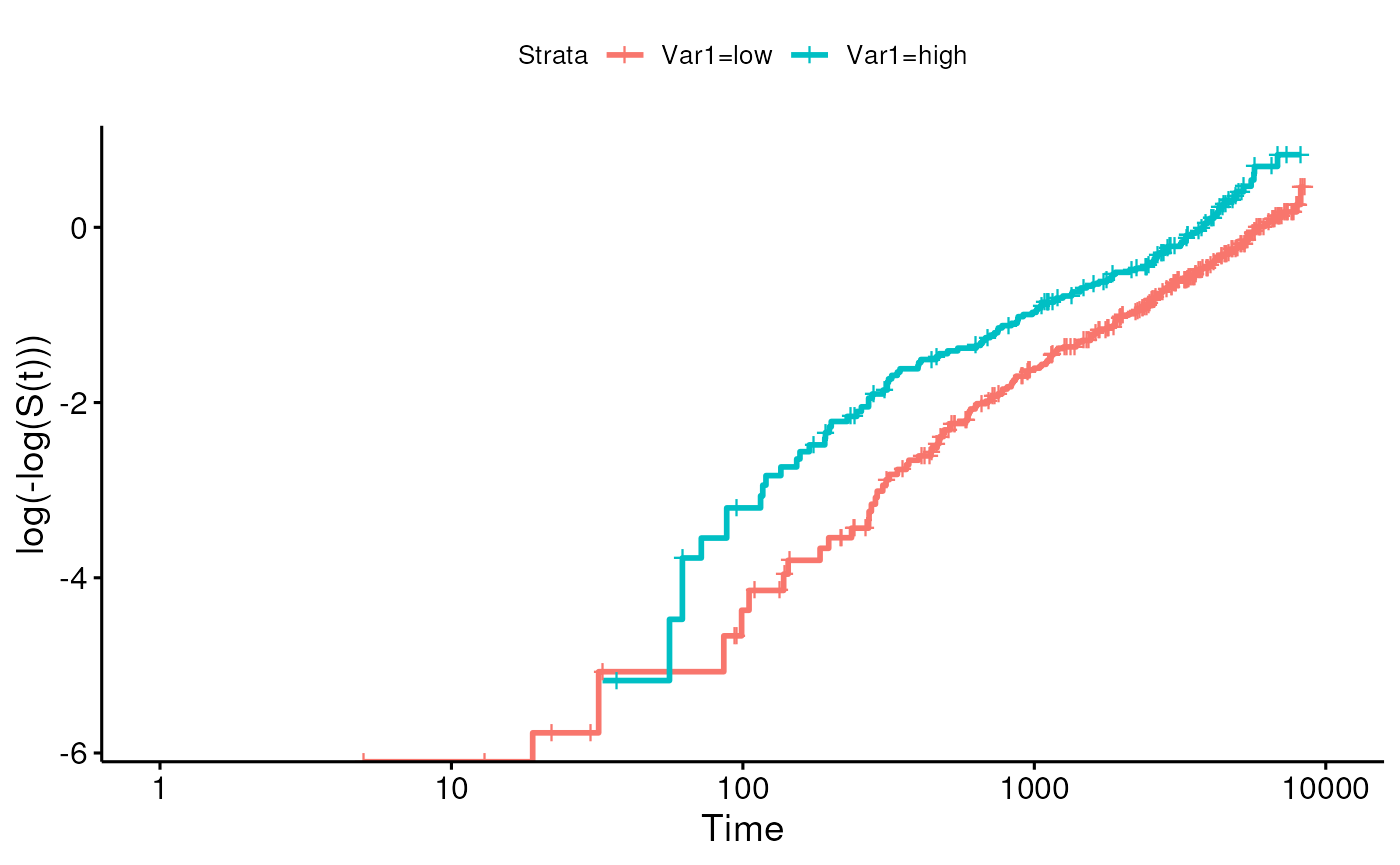
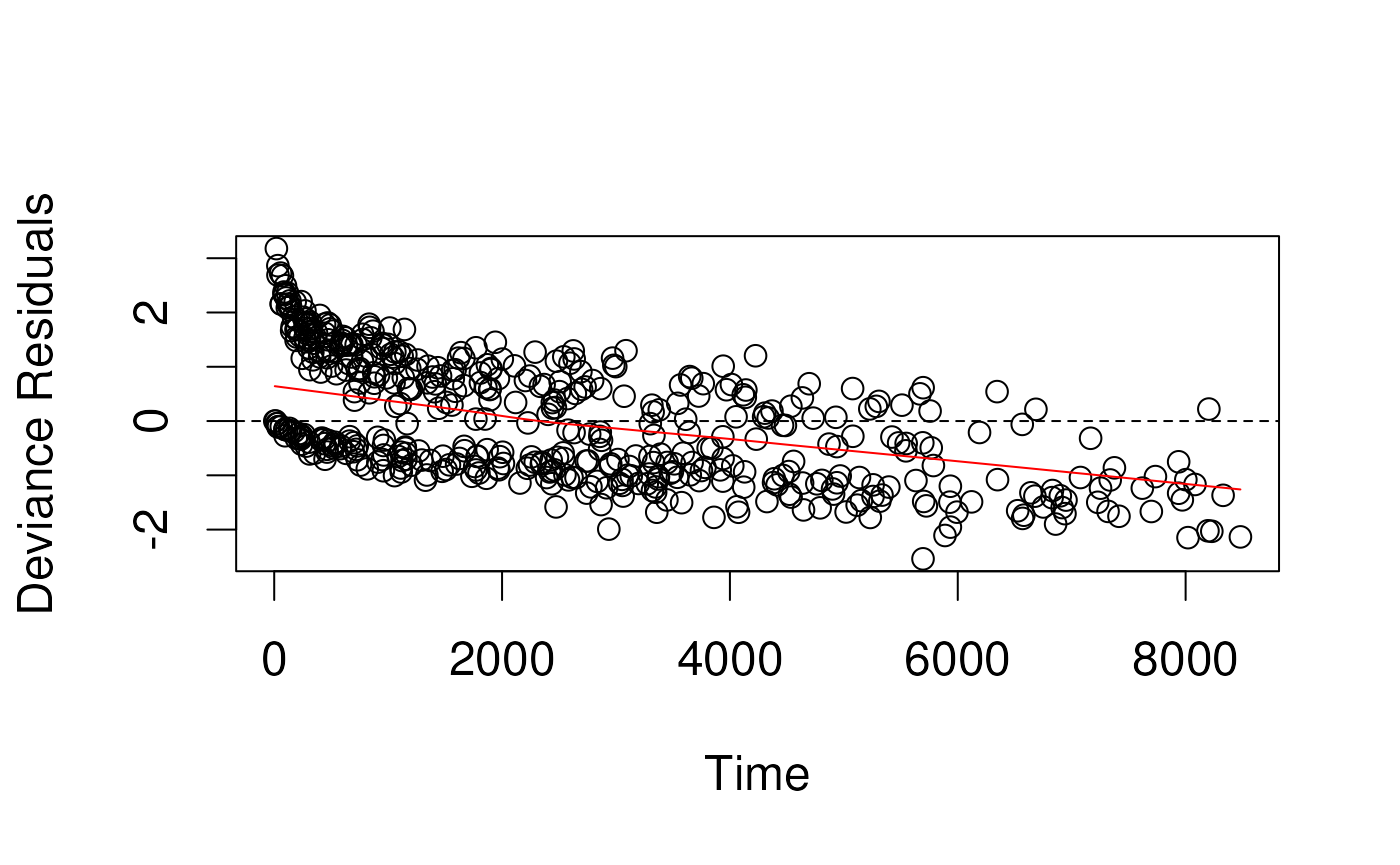
Deviance residuals in simulated data
- Deviance residuals are scaled Martingale residuals
- Should be more symmetrically distributed about zero?
- Observations with large deviance residuals are poorly predicted by
the model
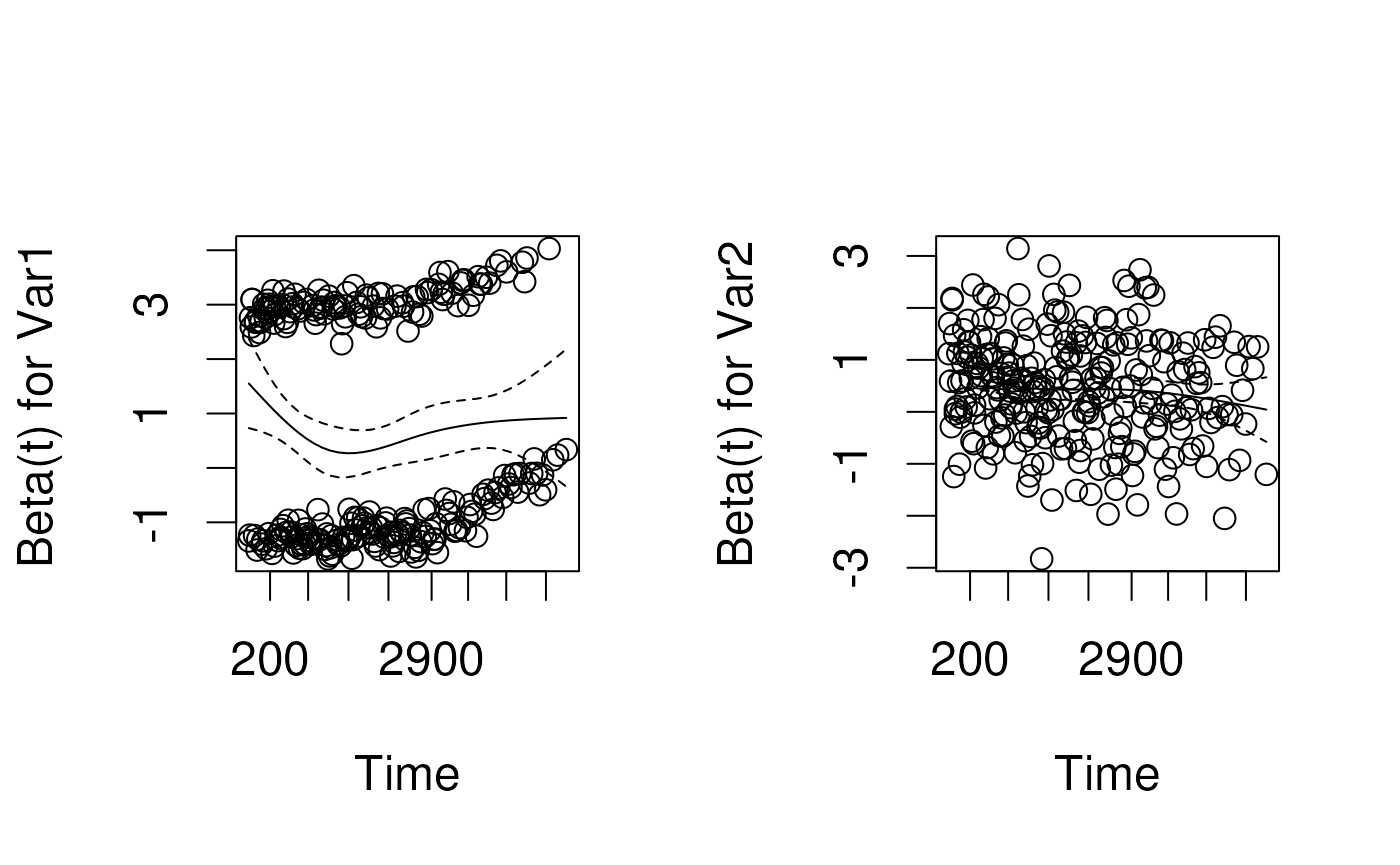
Schoenfeld residuals
- technical definition: contribution of a covariate at each event time to the partial derivative of the log-likelihood
- intuitive interpretation: the observed minus the expected values of the covariates at each event time.
- a random (unsystematic) pattern across event times gives evidence the covariate effect is not changing with respect to time
- If it is systematic, it suggests that as time passes, the covariate effect is changing.
Schoenfeld test for proportional hazards
- Tests correlation between scaled Schoenfeld residuals and time
- Equivalent to fitting a simple linear regression model with time as the predictor and residuals as the outcome
- Parametric analog of smoothing the residuals against time using LOWESS
- If the hazard ratio is constant, correlation should be zero.
- Positive values of the correlation suggest that the log-hazard ratio increases with time.
## chisq df p
## Var1 0.00887 1 0.925
## Var2 4.92734 1 0.026
## GLOBAL 5.07415 2 0.079Example: Primary Biliary Cirrhosis (PBC)
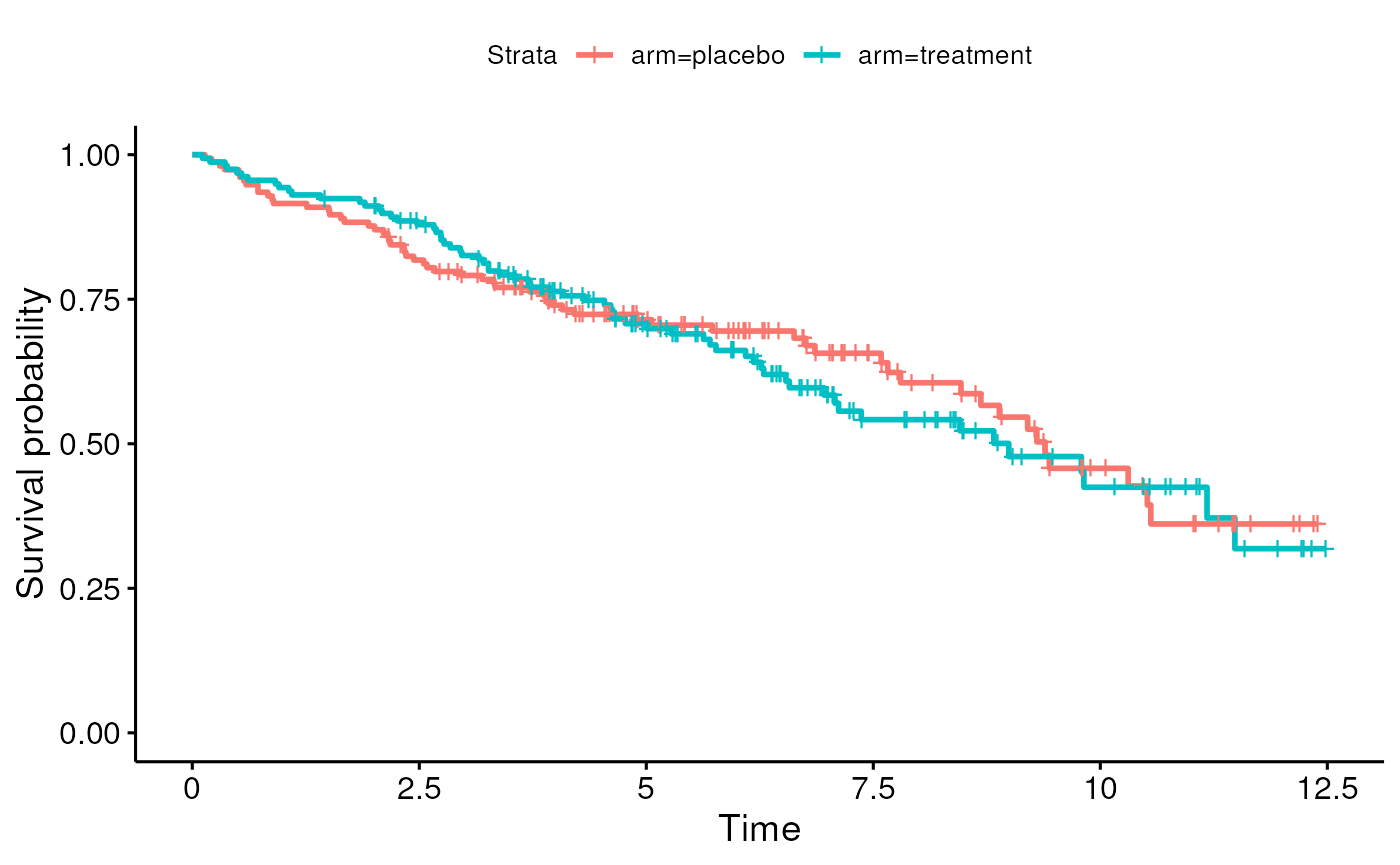
- Mayo Clinic trial in primary biliary cirrhosis (PBC) of the liver conducted between 1974 and 1984, n=424 patients.
- randomized placebo controlled trial of the drug D-penicillamine.
- 312 cases from RCT, plus additional 112 not from RCT.
- Primary outcome is (censored) time to death
Tests for trend
What are tests for trend?
- For models including an ordinal variablepush
- Such as PBC stage (1, 2, 3, 4), age category, …
- Is there a linear / quadratic / cubic relationship between coefficients and their order?
- Test by LRT or Wald Test
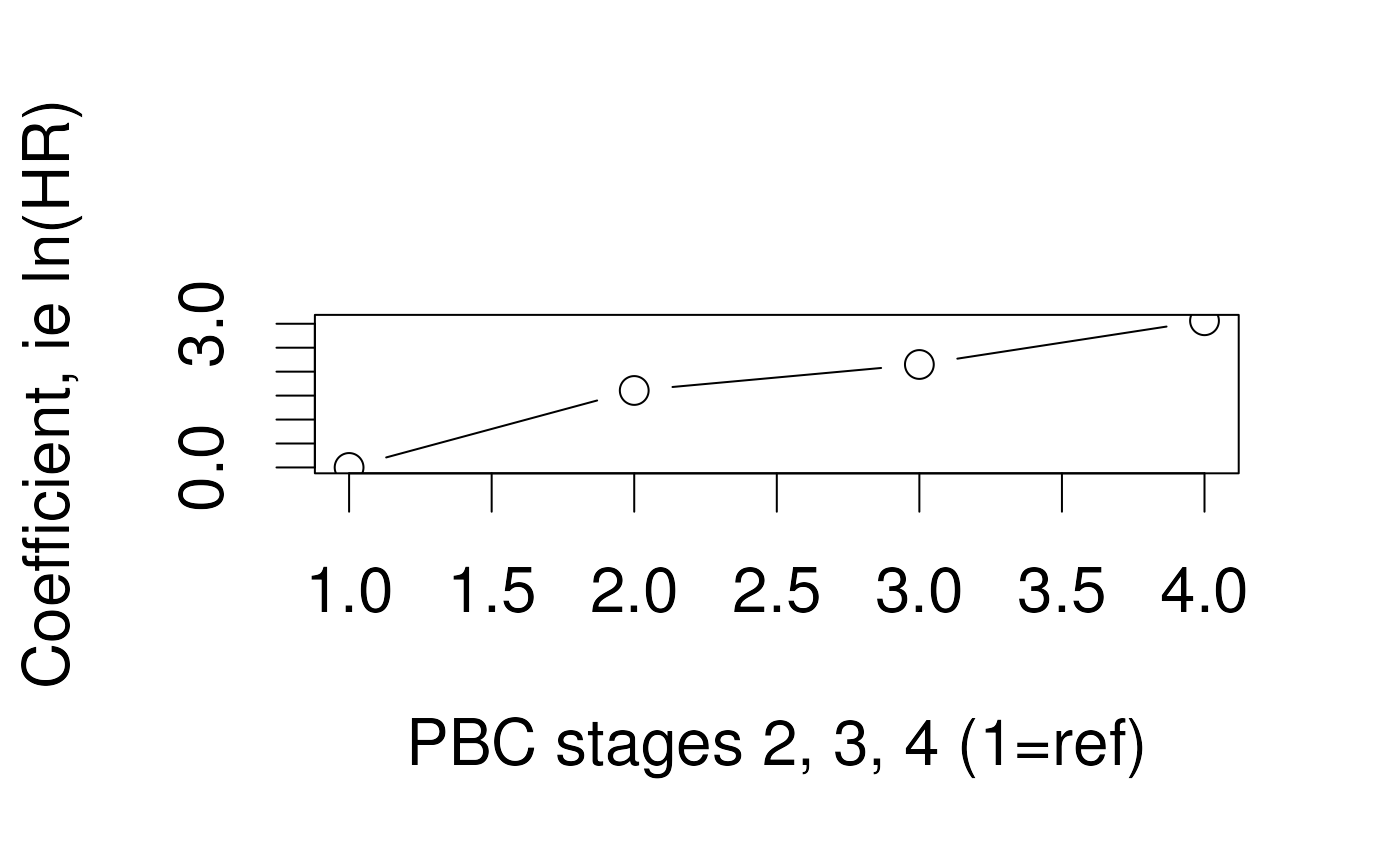
Fitting a test for trend in R
- Just define stage as an ordered factor and tests for trend are done automatically:
pbc.os <-
mutate(pbc.os, stageordered = factor(stage, ordered = TRUE))
fit <- coxph(Surv(time, os) ~ stageordered, data = pbc.os)
summary(fit) ## Call:
## coxph(formula = Surv(time, os) ~ stageordered, data = pbc.os)
##
## n= 312, number of events= 125
##
## coef exp(coef) se(coef) z Pr(>|z|)
## stageordered.L 2.1759 8.8099 0.6801 3.199 0.00138 **
## stageordered.Q -0.3469 0.7069 0.5248 -0.661 0.50867
## stageordered.C 0.3209 1.3784 0.2990 1.073 0.28316
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## stageordered.L 8.8099 0.1135 2.3231 33.410
## stageordered.Q 0.7069 1.4146 0.2527 1.977
## stageordered.C 1.3784 0.7255 0.7671 2.477
##
## Concordance= 0.702 (se = 0.022 )
## Likelihood ratio test= 52.74 on 3 df, p=2e-11
## Wald test = 43.92 on 3 df, p=2e-09
## Score (logrank) test = 53.85 on 3 df, p=1e-11Highly significant tests of overall fit by LRT, Wald, and logrank test.
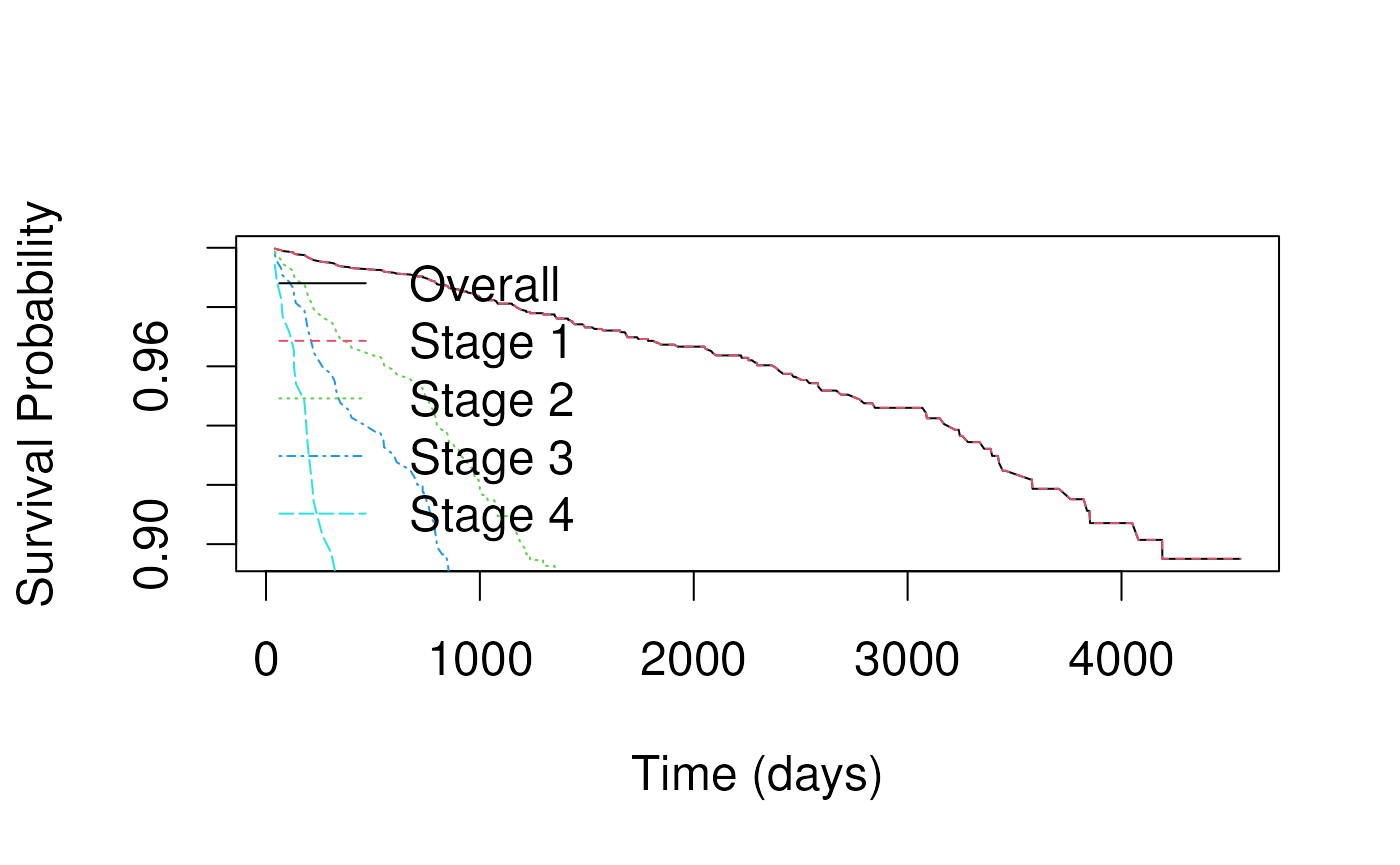
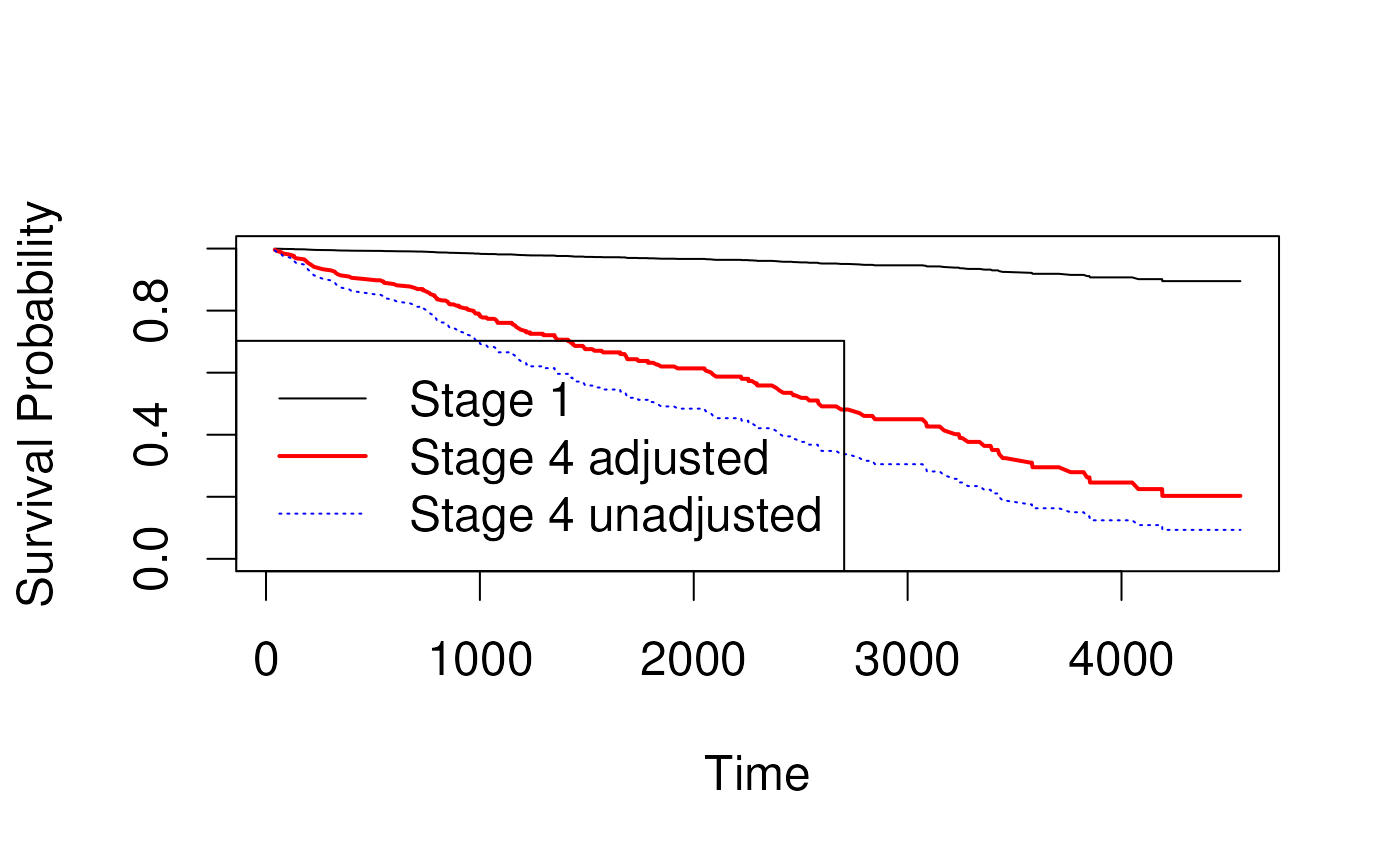
Predictions for specific covariate patterns
How to predict survival from a Cox model?
- The Cox model is a relative risk model
- only predicts relative risks between pairs of subjects
- Key is to calculate the overall , then multiply it by the relative hazard for the specific covariate pattern.
- In this example we plot the baseline survival for all stages together, then for stages 1-4 separately.
## Call:
## coxph(formula = Surv(time, os) ~ age + sex + edema + stage +
## arm, data = pbc.os)
##
## n= 312, number of events= 125
##
## coef exp(coef) se(coef) z Pr(>|z|)
## age 0.027618 1.028003 0.009362 2.950 0.00318 **
## sexf -0.317540 0.727938 0.248839 -1.276 0.20193
## edema0.5 0.538715 1.713804 0.275287 1.957 0.05036 .
## edema1 2.080422 8.007845 0.276959 7.512 5.84e-14 ***
## stage2 1.535263 4.642546 1.034854 1.484 0.13793
## stage3 1.998217 7.375893 1.016097 1.967 0.04923 *
## stage4 2.666263 14.386101 1.016234 2.624 0.00870 **
## armtreatment 0.057946 1.059658 0.189200 0.306 0.75940
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## age 1.0280 0.97276 1.0093 1.047
## sexf 0.7279 1.37374 0.4470 1.186
## edema0.5 1.7138 0.58350 0.9992 2.940
## edema1 8.0078 0.12488 4.6534 13.780
## stage2 4.6425 0.21540 0.6108 35.288
## stage3 7.3759 0.13558 1.0067 54.040
## stage4 14.3861 0.06951 1.9630 105.430
## armtreatment 1.0597 0.94370 0.7313 1.535
##
## Concordance= 0.77 (se = 0.022 )
## Likelihood ratio test= 107.6 on 8 df, p=<2e-16
## Wald test = 120.8 on 8 df, p=<2e-16
## Score (logrank) test = 177.1 on 8 df, p=<2e-16Predicted survival for adjusted coefficients
- Can create Kaplan-Meier curves for crude or unadjusted coefficients
- Section 6.3.2.3 in Vittinghoff
- Idea is to estimate hazard ratio in an unadjusted model:
## stage2 stage3 stage4
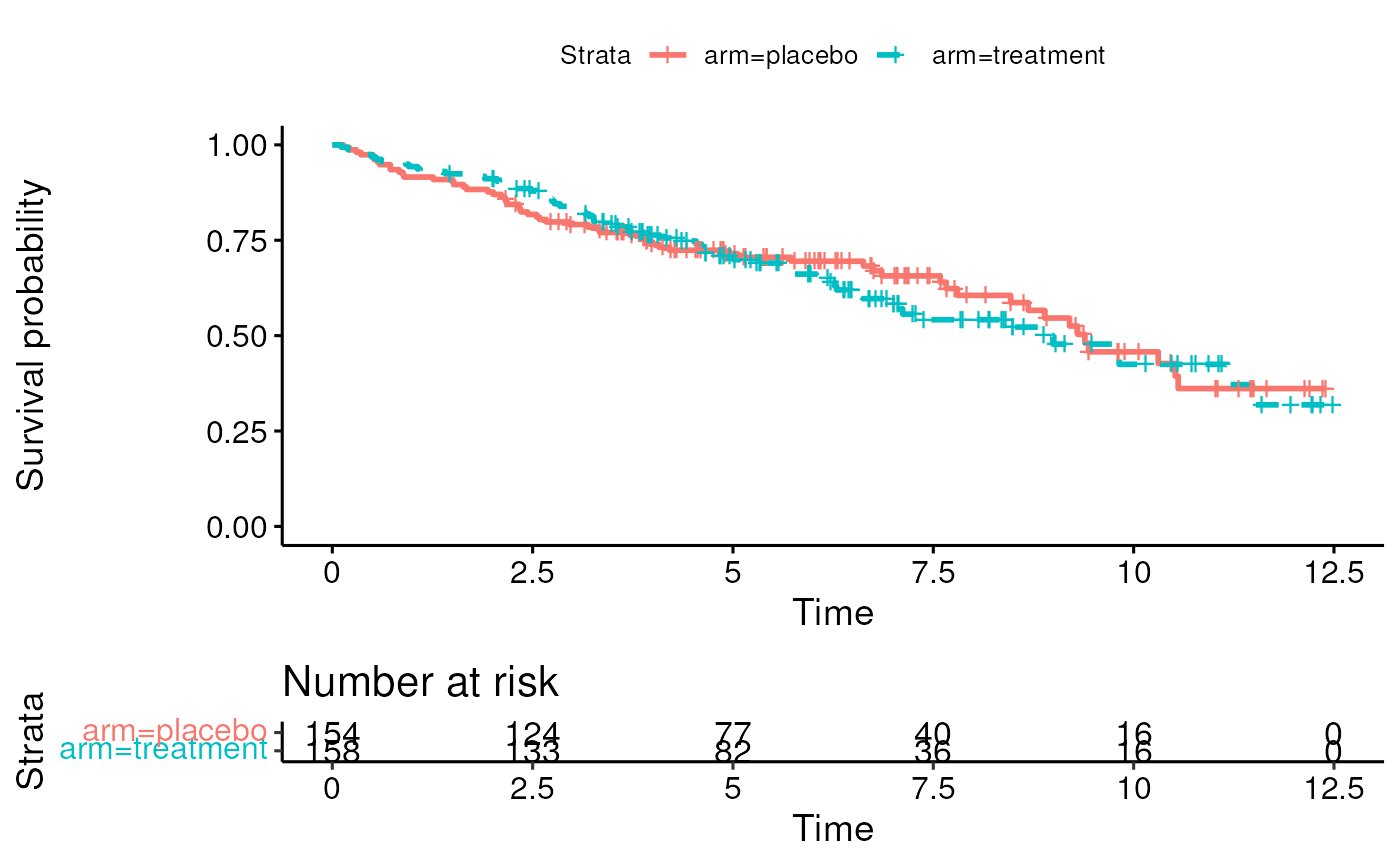
## 1.607014 2.149500 3.062775Stratification
What is stratification?
relevant to all kinds of regression, not just survival analysis
-
when analysis is separated into groups or strata
- must have an adequate number of events in each stratum (at least 5 to 7)
- can be used to adjust for variables with strong impact on survival
- can help solve proportional hazards violations
Strata have different baseline hazards
Coefficients / Hazard Ratios are calculated within stratum then combined.
Vittinghoff 6.3.2
How to stratify
Example - in R, strata() can be added to any model formula
## Call:
## coxph(formula = Surv(time, os) ~ trt + strata(stage), data = pbc.os)
##
## n= 312, number of events= 125
##
## coef exp(coef) se(coef) z Pr(>|z|)
## trt -0.1063 0.8992 0.1814 -0.586 0.558
##
## exp(coef) exp(-coef) lower .95 upper .95
## trt 0.8992 1.112 0.6302 1.283
##
## Concordance= 0.494 (se = 0.025 )
## Likelihood ratio test= 0.34 on 1 df, p=0.6
## Wald test = 0.34 on 1 df, p=0.6
## Score (logrank) test = 0.34 on 1 df, p=0.6Competing Risks Data
What are competing risks?
- Example from Vittinghoff 6.5: The MrOS study (Orwoll et al. 2005) followed men over 65 to examine predictors of bone fracture and low BMD (subclinical bone loss)
- At end of study participants had:
- developed fracture (outcome of interest),
- remained alive without fracture (incomplete follow-up), or
- died prior to fracture (incomplete follow-up)
Orwoll, E. et al. (2005). Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study–a large observational study of the determinants of fracture in older men. Contemporary Clinical Trials, 26(5), 569–585.
Why not treat died prior to fracture and alive without fracture as censored?
- Recall the independent censoring assumption (Vittinghoff 6.6.4):
- censored people are similar to those who remain at risk in terms of developing the event of interest;
- censoring is independent of the event of interest.
- For patients who died this assumption is highly suspect
Reasons for right censored data
- Cut-off date of analysis (administrative censoring):
- Censoring usually independent
- Loss to follow-up
- Independence may be problematic if sicker individuals discontinue participant in study (lack of energy, too ill, return to home country)
- or if healthier individuals discontinue participation (don’t feel the need to continue, start new life in other country)
- Competing risks:
- Often informative.
- In competing risks analysis, independence between competing risks is not required
Very brief summary of competing risk methods
- 1-to-1 mapping between hazard and cumulative incidence function is lost in competing risks
- Standard Kaplan-Meier estimator is biased for competing risks data
- Aalen-Johansen estimator is better choice
- Gary’s test is analogous to log-rank test
- cause-specific standard Cox PH model might be useful for prognostic (causal) testing, but not estimating a population Hazard Ratio
Resources for competing risk methods
- Z. Zhang, Survival analysis in the presence of competing risks, Ann Transl Med. 2017 Feb; 5(3): 47. PMID: 28251126
- cmprsk package
- riskRegression package
Propensity score analysis
What is propensity score analysis?
- an alternative to multivariate regression to control for hypothesized confounders in observational studies:
outcome ~ exposure + counfounder1 + confounder2- a stratification approach that is more practical than stratifying on multiple hypothesized confounders
- an approach to summarizing many covariates into a single score
- a convenient approach to controlling for many hypothesized confounders
Propensity score approach to correction for confounders
- Step 1: fit the propensity score model (no outcome) that predicts propensity for exposure based on confounders:
exposure ~ counfounder1 + confounder2Step 2: use propensity predictions to match or stratify participants with similar propensity (for example, stratifying on quintiles of propensity)
Step 3: check adequacy of matching or stratification, ie by comparing attributes of matched participants
Step 4: test hypothesis among matched participants:
outcome ~ exposurePropensity score references
- P.C. Austin (2011), An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies. Multivariate Behavioral Research, 46:3, 399-424, DOI: 10.1080/00273171.2011.568786
- R. d’Agostino (1998), Tutorial in Biostatistics: propensity score methods for bias reduction in the comparison of a treatment to a non-randomized control group. Stat. Med. 17, 2265-2281. http://www.stat.ubc.ca/~john/papers/DAgostinoSIM1998.pdf
- You don’t need any special package to do basic propensity score matching (e.g. stratifying by quintiles), but the MatchIt package provides multiple matching approaches, diagnostics, good documentation