Session 2: Linear and logistic regression as Generalized Linear Models
Levi Waldron
session_lecture.RmdLearning Objectives and Outline
Review of multiple linear regression
Linear Regression as a GLM
Generalized Linear Models (GLM)
- Linear regression is a special case of a broad family of models called “Generalized Linear Models” (GLM)
- This unifying approach allows to fit a large set of models using maximum likelihood estimation methods (MLE) (Nelder & Wedderburn, 1972)
- Can model many types of data directly using appropriate distributions, e.g. Poisson distribution for count data
- Transformations of not needed
Components of GLM
-
Random component specifies the conditional
distribution for the response variable
- doesn’t have to be normal
- can be any distribution in the “exponential” family of distributions
- Systematic component specifies linear function of predictors (linear predictor)
-
Link [denoted by g(.)] specifies the relationship
between the expected value of the random component and the systematic
component
- can be linear or nonlinear
Logistic Regression as a GLM
The logistic regression model
- The model:
Random component: follows a Binomial distribution (outcome is a binary variable)
Systematic component: linear predictor
Link function: logit (log of the odds that the event occurs)
The logit function
logit <- function(P) log(P/(1-P))
plot(logit, xlab="Probability", ylab="Log-odds",
cex.lab=1.5, cex.axis=1.5)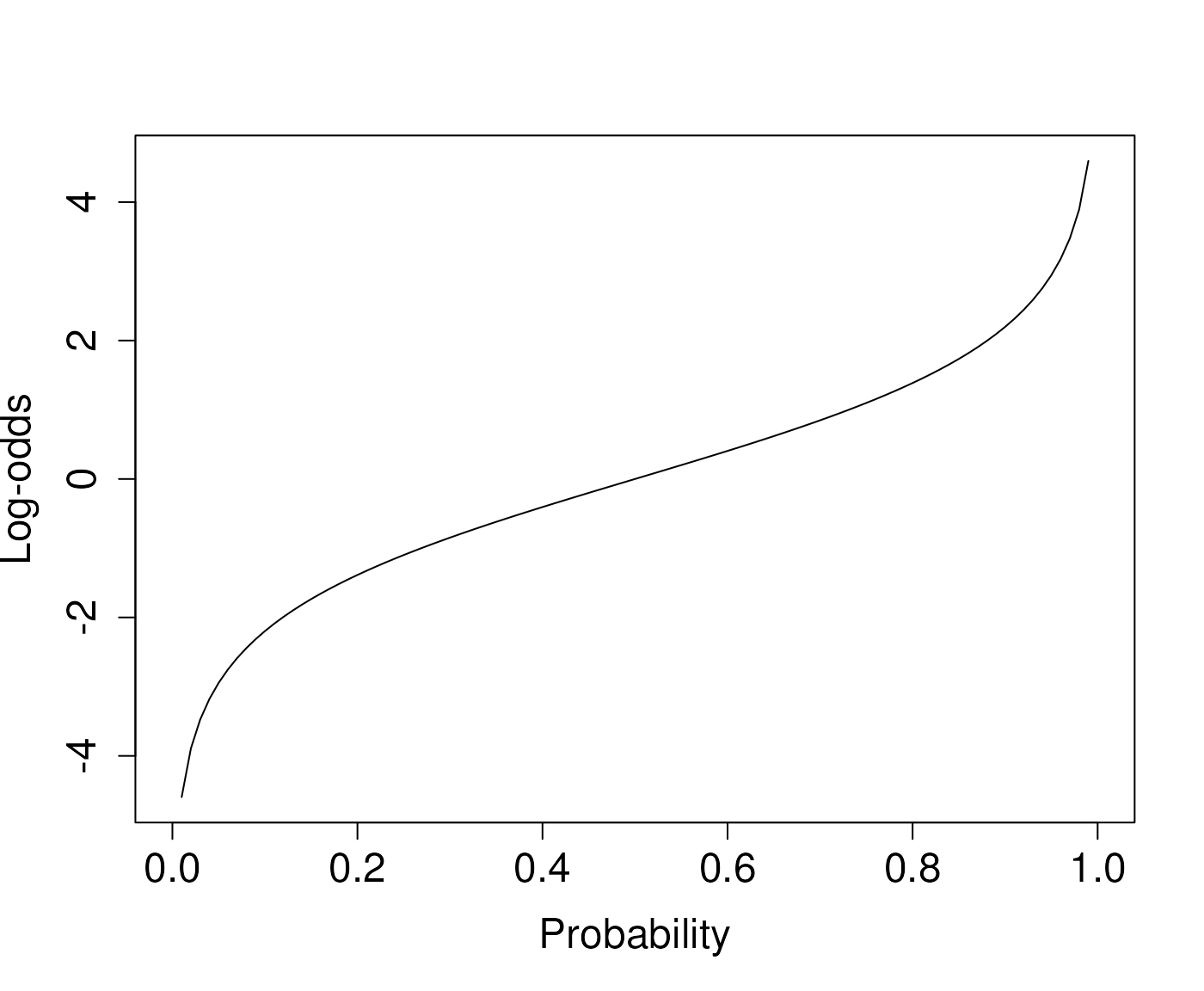
Example: contraceptive use data
| Age (years) | <25 (N=4) |
25-29 (N=4) |
30-39 (N=4) |
40-49 (N=4) |
Overall (N=16) |
|---|---|---|---|---|---|
| education | |||||
| high | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 8 (50.0%) |
| low | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 8 (50.0%) |
| wantsMore | |||||
| no | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 8 (50.0%) |
| yes | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 2 (50.0%) | 8 (50.0%) |
| percentusing | |||||
| Mean (SD) | 18.8 (7.64) | 27.1 (6.53) | 38.8 (15.6) | 46.9 (23.8) | 32.9 (17.5) |
| Median [Min, Max] | 18.2 [10.2, 28.6] | 27.6 [18.9, 34.5] | 39.5 [22.8, 53.4] | 50.5 [14.6, 72.1] | 28.3 [10.2, 72.1] |
Source: http://data.princeton.edu/wws509/datasets/#cuse. Note, this table represents rows of the source data, not number of participants. See the lab to make a table that summarizes the participants.
Perform regression
- Outcome: whether using contraceptives or not
- Predictors: age, education level (high/low), whether wants more children or not
fit1 <- glm(cbind(using, notUsing) ~ age + education + wantsMore,
data=cuse, family=binomial("logit"))
summary(fit1)##
## Call:
## glm(formula = cbind(using, notUsing) ~ age + education + wantsMore,
## family = binomial("logit"), data = cuse)
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -0.8082 0.1590 -5.083 3.71e-07 ***
## age25-29 0.3894 0.1759 2.214 0.02681 *
## age30-39 0.9086 0.1646 5.519 3.40e-08 ***
## age40-49 1.1892 0.2144 5.546 2.92e-08 ***
## educationlow -0.3250 0.1240 -2.620 0.00879 **
## wantsMoreyes -0.8330 0.1175 -7.091 1.33e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 165.772 on 15 degrees of freedom
## Residual deviance: 29.917 on 10 degrees of freedom
## AIC: 113.43
##
## Number of Fisher Scoring iterations: 4Residuals for logistic regression
Pearson residuals for logistic regression
- Traditional residuals don’t make sense for binary .
- One alternative is Pearson residuals
- take the difference between observed and fitted values (on probability scale 0-1), and divide by the standard deviation of the observed value.
- Let be the best-fit predicted probability for each data point, i.e.
- is the observed value, either 0 or 1.
Summing the squared Pearson residuals produces the Pearson Chi-squared statistic:
Deviance residuals for logistic regression
- Deviance residuals and Pearson residuals converge for high degrees of freedom
- Deviance residuals indicate the contribution of each point to the model likelihood
- Definition of deviance residuals:
Where if and if .
- Summing the deviances gives the overall deviance:
Likelihood and hypothesis testing
What is likelihood?
- The likelihood of a model is the probability of the
observed outcomes given the model, sometimes written as:
- .
- Deviance residuals and the difference in log-likelihood between two models are related by:
Likelihood Ratio Test
- Use to assess whether the reduction in deviance provided by a more complicated model indicates a better fit
- It is equivalent of the nested Analysis of Variance is a nested Analysis of Deviance
- The difference in deviance under is chi-square distributed, with df equal to the difference in df of the two models.
Likelihood Ratio Test (cont’d)
fit0 <- glm(cbind(using, notUsing) ~ -1, data=cuse,
family=binomial("logit"))
anova(fit0, fit1, test="LRT")## Analysis of Deviance Table
##
## Model 1: cbind(using, notUsing) ~ -1
## Model 2: cbind(using, notUsing) ~ age + education + wantsMore
## Resid. Df Resid. Dev Df Deviance Pr(>Chi)
## 1 16 389.85
## 2 10 29.92 6 359.94 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1